The work program consists of six projects grouped into three themes that pursue the Lifespan AI vision from different perspectives: Data and Methods (D), Models and Interpretation (M), and Inference and Causality (C). D1 will advance DL strategies to explore and process long-term temporal change based on integration of high-dimensional data from multiple sources; D2 will combine neural networks and mixed-effects models to predict individual health trajectories over the life course; M1 will develop “normalizing flow” methods to derive joint distributions and conditional densities for health data; M2 will create a cognitive digital twin from everyday human activities to predict change across age groups; C1 will develop time-adaptive, explainable AI methods for recurrent neural networks and event times; and C2 will derive a framework for “causal discovery” in longitudinal studies, combining different data sets and accounting for nonlinearities.
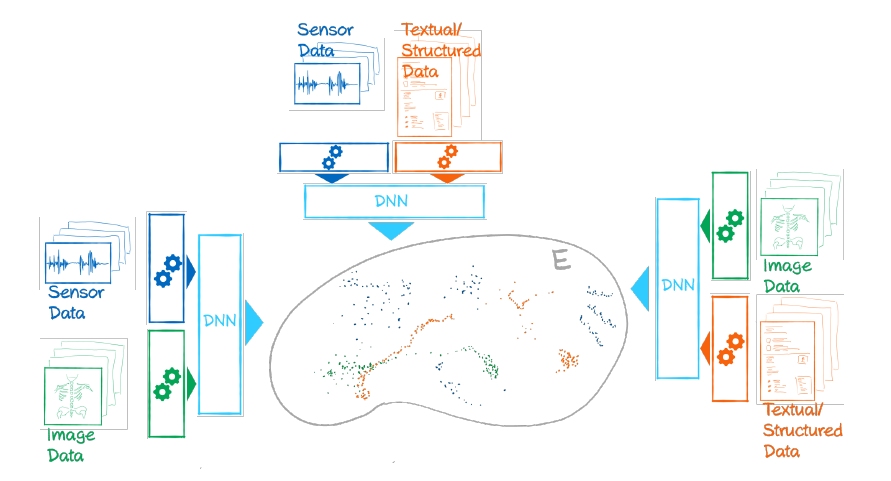
Long Delta Health Data encompass the entire life of individuals and cohorts. They stem from different and changing sources and are typically sparse and variable. The different sources, called modalities, and their varying temporal resolution even within one study hamper computational integration substantially, and between different studies with only partially shared modalities and partial temporal overlap, this obstacle gets even worse. To integrate such data, and make them amenable for computational analysis, the project will hence have to overcome these challenges. To devise integration methods with a high level of generalisation and applicability, we propose to employ deep-learning-based models. We will employ learnable and transferable data embedding models for time-resolved units comprising one or more modalities. Conceptually, this approach builds on the so-called attention modules known from language transformer models. The resulting embeddings will need to be general, robust, and amenable to “transfer learning”-like approaches, and we will validate the success with the unique datasets available to the project. In our project, we use datasets from two large German population studies that span a wide range of temporal resolutions and data types, such as questionnaires, voice recordings, and medical images. We aim to build a library of trained embedding models that can be used for ”new” data beyond the three development datasets. Further, on selected use cases, we will demonstrate how to use embedded data for predictive tasks. Predictions may encompass the imputation of missing data with regard to modalities, the interpolation of data between measured time points, or the extrapolation of one or more modalities into the future. Reliable predictions will depend on a certain structure of embedding space. Consequently, an important scientific project goal is to develop meaningful metrics to characterise embedding spaces quantitatively with regard to data quality issues, outliers, bias, and inconsistencies limiting the predictive capacity. The results obtained in this project have the potential to profoundly impact data science with a particular focus on the health domain, where time resolved data with a huge variability of data types and quality levels are predominant. As our aim is to devise concepts to unify vastly different data types and information sources with long delta characteristics, we believe the results will be applicable to areas even beyond health sciences.
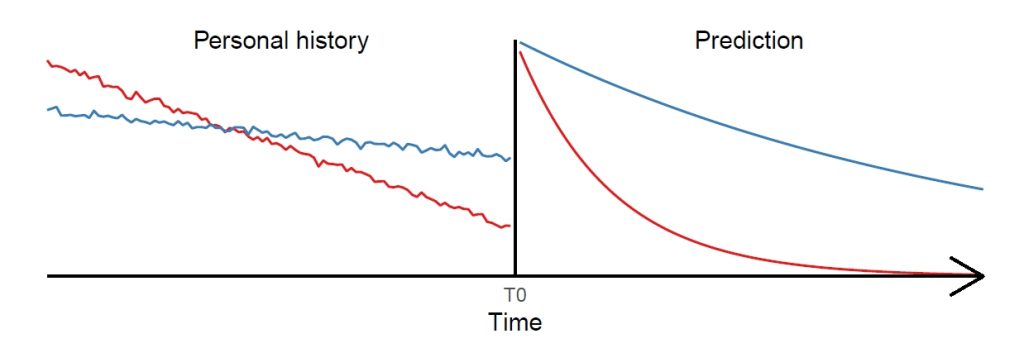
Chronic diseases such as obesity, cardiovascular diseases or dementia typically evolve over long time spans before becoming manifest and some were suggested to have their origin even in utero. Studying the developmental mechanisms of diseases with long latency and predicting (markers of) diseases long before an outbreak is very challenging. As there is almost no study covering the total lifespan, the joined analysis of multiple cohorts covering different periods in life provides the most promising approach to gain insights into long-term disease processes. So far, there is a striking lack of methods allowing to adequately analyse and predict the complex interplay of various factors based on pooled data of multiple cohorts. The proposed project intends to fill this gap by developing lifespan artificial intelligence (AI) methods suited to the prediction of individuallevel health trajectories over extended time spans. Generalised linear mixed-effects models (GLMM) display a flexible tool commonly used in epidemiology for modelling longitudinal and clustered data. However, among other drawbacks they are based on restrictive parametric assumptions. Flexible nonlinear machine learning (ML) methods like random forests (RF) and deep neural networks (DNN) mitigate the limitations but implicitly assume the data to be independent and identically distributed leading to inefficient estimates in a longitudinal setting. To combine their particular strengths while mitigating limitations, in this project we aim to advance so-called mixed-effects machine learning (ME-ML) approaches. In particular, we will advance ME-ML approaches for the prediction of individual health trajectories based on pooled cohort data integrating the random-effects structure of GLMM into RF and DNN, and assess the time span that can be validly predicted beyond the actual measurement period based on the devised method. We will further study how to best integrate data of multiple cohorts to generate a harmonised dataset as well as how the design features of multi-cohort studies such as the periods of overlap between cohorts affect the identifiability and performance of standard statistical methods for lifespan predictions and causal discovery. In summary, theoretical and practical investigations with regard to data harmonisation and the design features of multi-cohort studies will be complemented by methodological developments where promising candidates for lifespan AI methods based on ME-ML will be advanced and validated. Finally, the predictive performance of the newly developed methods will be compared with standard methods and evaluated considering different potential study designs and statistical issues in multi-cohort studies. Data from several cohorts will be used for illustration and validation of methods.
Health sciences use statistical models to quantify health status and outcome as it evolves over time and is influenced by risk factors and/or treatments. Hereby the quantification of uncertainty is a crucial methodological contribution of statistics. Since trend and uncertainty are best described in terms of distributions, a model for the joint distribution of all variables and features over all time points under consideration provides the most complete statistical picture. This permits – at least formally – all conclusions statistics can provide without untestable assumptions. Such an approach is not straightforward and requires machine learning methods when the data are high dimensional. Further challenges arise in lifespan data, where the data are of different scales, are measured at individual specific time points and come from different data sources with only partial overlap or from study cohorts whose variable sets change over time. In this project, we tackle these challenges by the development of normalizing flows that utilise invertible residual neural networks and use generalised linear mixed models (GLMM) for the base distributions. The use of GLMMs is particularly attractive for health scientists because they are frequently used in applications. Normalizing flows based on invertible residual networks have the major advantage of providing analytical expressions for the joint distribution that can be well utilised for the anticipated statistical and scientific conclusions. The key idea of our method is to start modelling the conditional distribution of each variable given the other variables by nonlinear transformations of GLMMs and to combine them to an global joint distribution by an average of randomly selected sequential factorisations which can be achieved by sequentially reducing the set of variables we condition on in the conditional distributions (“reverse marginalisations”). After deriving an approach to fit a joint model with complete observations, we will develop an algorithm for updating the model with incomplete observations and extending it by additional variables. The joint distribution will then be utilised to obtain (overall regularised) estimates of scientifically interesting conditional distributions from which point and interval predications can be derived. We will also develop methods to interpret the overall (black box) model and to investigate its internal and external validity. While the joint model already accounts for aleatoric uncertainty (by modelling distributions) we will work out methods to account also for epistemic uncertainty, i.e. approaches for quantifying the model fit and to account for the model uncertainty in prediction intervals. The new methods will be applied to and illustrated with data and variables from the IDEFICS/I.Family cohort, the NAKO Health Study and GePaRD data which are all collected and/or managed by BIPS.
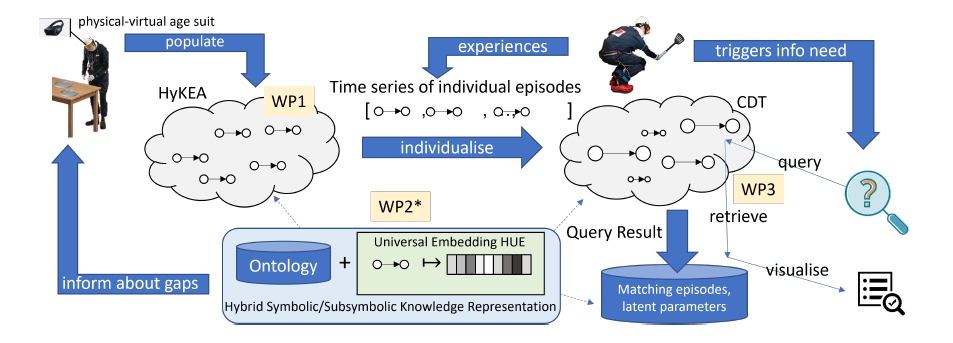
This project investigates computational methods for representation, reasoning, and modelling of capabilities and impairments, causally related to diseases, of individuals performing everyday activities. The goal is to make predictions about whether individuals need support and for what, how their impairments progress over time, and how this depends on contextual factors, like the structuring of the environment or fatigue. To this end, our project will design, realise, and examine the computational principles of a Lifespan Predictive Modelling Engine (L-PME), a hybrid knowledge representation and reasoning framework that accompanies individuals, records their everyday activities, and maintains individual models of impairments in a lifelong learning process. To represent an individual person, L-PME customises a cognitive digital twin from a generic impairment-aware knowledge representation of everyday activities that captures (1) the development of disease-induced impairments in context of a person’s changing situations, needs, and tasks, (2) the causal influence of capabilities and impairments on everyday manipulation tasks, and (3) the latent interdependence of behaviours in different everyday activities to answer open queries and predict performance and behaviour. We will investigate the foundations of the representation, reasoning, and modelling methods by applying it to persons wearing physical-virtual age suits during everyday activities, such as setting the table. This allows us to investigate the principles of L-PME under observable and controllable conditions by changing the parameters of the physical-virtual age suits systematically in an expert-in-the-loop process.
The recent rise of artificial intelligence (AI) in almost all research fields is closely followed by methods of explainable AI (XAI), which try to explain AI systems and black box machine learning models. These methods proved to perform well in many research fields, including applications on health data. For example, XAI methods are used to detect relevant pixel regions in medical imaging. However, in lifespan health data, we face heterogeneous input data with varying data structures and dimensions, longitudinal predictors measured over long periods of time and time-dependent outcomes. It is particularly challenging to integrate such data into AI systems and it is equally challenging to explain or interpret the resulting models. In this project, we tackle this challenge by developing XAI methods that can handle data covering the whole lifespan, including longitudinal predictors, time-dependent outcomes and combinations of different data sources, which change over the lifespan. We address both interpretation, i.e. understanding a model by itself, and explanation, i.e. using post-hoc methods on already trained models. For interpretation, we use invertible neural network structures, which directly relate the network’s output with its input. For explanation, we develop methods based on attributions (e.g. gradient-based), attention mechanisms and lossbased methods. Further, we develop methods to generate so-called knockoffs for lifespan health data with normalizing flows, aiming at high dimensional conditional variable selection and conditional independence testing on such complex data. In summary, we aim at explaining and interpreting the AI models developed in the research unit. That is crucial for the research unit and, more generally, for health science because it contributes to the major goal of understanding health and disease mechanisms. It is also important for the general field of AI because our methodological advancements generalise beyond health data, e.g. to text or speech processing.
The project will capitalise on AI methods to develop novel robust approaches for the discovery and characterisation of causal relations across the lifespan. Such methods are urgently needed because randomised trials, the gold standard for inferring causality, are almost never available for questions regarding long-term effects, e.g. of lifestyle in early childhood, across the whole lifespan. Causal discovery comprises a class of methods whereby a multivariate dataset is taken as input and, ideally, the output is a directed acyclic graph encoding the probabilistic causal structure among variables (nodes). While such methods are slowly being adopted in the biomedical and social sciences, they have never been applied to the case where several different datasets with temporal structure are combined to cover a long range of a person’s lifespan – our proposal aims to fill this gap. The project will begin with laying the foundation by delivering a theoretically sound and practically useful discovery algorithm which combines time-structured datasets allowing us to cover a long lifespan: the TIOD algorithm – “temporal integration of overlapping datasets”. To ensure that this algorithm can be applied to nonlinear / nonparametric settings we will exploit state-of-the-art machine learning methodologies, such as causal generative neural networks. A particular focus will also be given to the role of multicohort study designs for the performance of TIOD, as some designs may be more informative than others. Further, we will generalise methods for quantifying causal relations, such as those found by TIOD, so as to estimate long-term effects from separate data sources. To validate and demonstrate the practical usefulness of our methods we will elicit epidemiological research questions and prepare real datasets, combing for instance the IDEFICS/I.Family cohort study with the NAKO Health Study, where challenging issues of data harmonisation will need to be addressed. These applications to real data will ultimately deliver novel insights, for instance, into causal relations between early childhood factors and health outcomes in adulthood.
The University of Bremen provides two bridge positions for Lifespan AI.
One position for a discipline connector:
One position for a data steward:
